The whole-genome sequencing of two Pakistani novel coronaviruses (2019-nCoV) has revealed almost contrasting genomic features to each other when compared with the Chinese coronavirus reference sequence.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) isolate Wuhan-Hu-1, with accession number MN908947, was the first complete genome submitted to the NCBI database.
As of 03 April 2020, two coronavirus whole-genome sequences from Pakistan have been sequenced and deposited in GenBank. The first coronavirus genome (SARS-CoV-2/Gilgit1/human/2020/PAK) was submitted on 25 March 2020 by the Department of Healthcare Biotechnology, National University of Sciences and Technology (NUST), Islamabad-Pakistan. The second coronavirus genome (isolate SARS-CoV-2/Manga1/human/2020/PAK) was submitted 30 March 2020 by the University Institute of Biochemistry and Biotechnology, PMAS Arid Agriculture University, Rawalpindi-Pakistan.
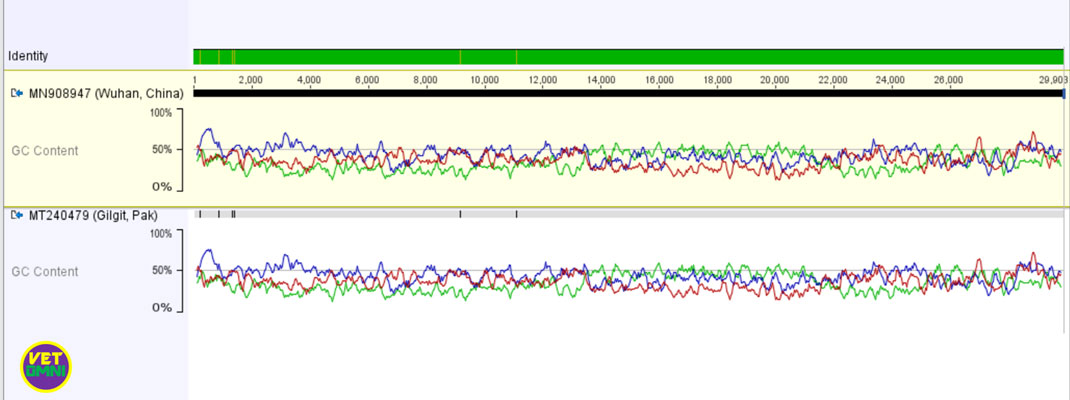
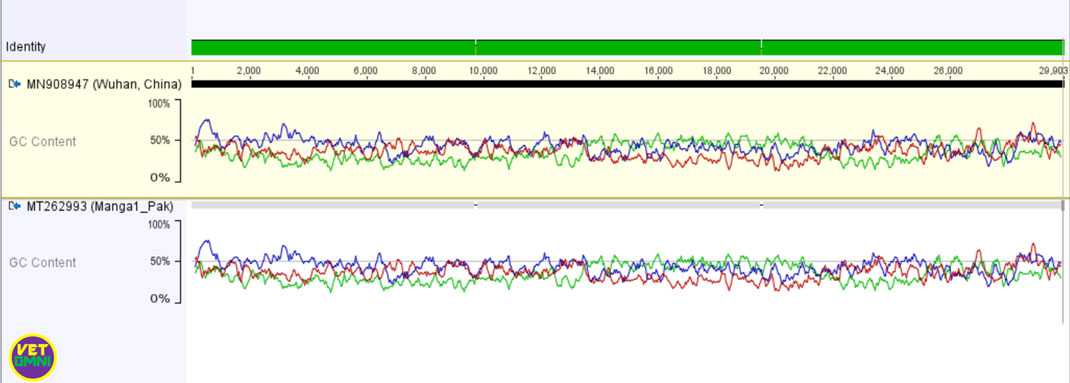
Pakistani isolate SARS-CoV-2 (accession number: MT240479) from Gilgit has a 99.98 % nucleotide identity with Chinse SARS-CoV-2 isolate while the other from Manga1 (accession number: MT262993) has almost 100% nucleotide identity except for two multinucleotide deletions.
When the nucleotide sequence of each Pakistani novel coronavirus (2019-nCoV) strain was compared to the Chinese sequence, the sequence from Gilgit, Pakistan revealed a total of 6 nucleotide mutations/substitutions. In the other sequence SARS-CoV-2/Manga1, Pakistan, two big nucleotide deletions were found.
“One multinucleotide deletion is of 15 bp, while the other is of 21 bp length,” says the bioinformatic analyst Dr. Muhammad Akbar Shahid, who is an Assistant Professor of Microbiology at Bahauddin Zakariya University, Multan-Pakistan.
He said, “The effect of these mutations/deletions on the pathogenicity and transmission potential of these Pakistani novel coronaviruses still needs to be explored and confirmed by the experiments.”
Novel coronavirus (2019-nCoV) that emerged in Chinese city Wuhan, and later, spread to more than 202 countries in almost all continents except Antarctica, has been sequenced by the researchers from different countries and almost 364 whole/partial genomes have been deposited in the GenBank so far.
Overall, a total of 277 whole/partial sequences (>29000 bp) have been deposited by the USA alone, the largest number among all countries so far.
As of 3 April 2020, a total of 68 SARS-CoV-2 sequences from Asia, 13 from Europe, 277 from North America, 1 from Australia, and 3 from South America have been sequenced and submitted to the GenBank. From the Asian continent, China (54), India (2), Japan (3), Nepal (1), Pakistan (2), South Korea (1), Taiwan (3), and Viet Nam (2) have deposited their SARS-CoV-2 sequences in the NCBI database.
At least 8 strains of the coronavirus are circulating in the world. Over 2000 genetic sequences have revealed that it is mutating on average every 15 days. According to the researchers, the virus is mutating so slowly that its strains are fundamentally very similar to each other. It is still not clear if these mutations are affecting its virulence, the ability to cause more damage.
Scientists say as the coronavirus is not mutating so quickly, it might respond to a single vaccine. Nucleotide sequence data suggests that the virus is genetically stable, which could increase the effectiveness of the current vaccines in development.
Author: Dr. Muhammad Akbar Shahid, Assistant Professor (Microbiology), Bahauddin Zakariya University, Multan-Pakistan
Very informative….but as it is RNA virus it can mutate easily….my question is can two strains co infect same person….if yes then it may develop new strains by genetic drift or shift????? Then how could single vaccine work????
I don’t think being an RNA virus makes it mutate easily; however, viruses do mutate naturally at some rate depending on their genetic nature. Yes, two strains can infect the same person as long as their ability to bind the receptors is not changed. As long as their pathogenicity determinants remain the same, one vaccine can work. The determination of genetic recombination events for viruses is an active area of research. When vaccines tend to become ineffective in the patients, then it’s time for scientists to start thinking to develop new vaccines. So, it all depends where the mutations are and what are their effects on the viral virulence/immunogenicity, ability to evade the host immune responses etc.
Great .
Inshallah pathogenicity factors will be determined soon and hope so vaccines will be developed .
Excellent Sir.Akbar Shahid
Good Work Sir…
Excellent Work Sir
Good effort Dr. Sb.
Stupendous???
Ma Sha Allah excellent work for Nation and proud moment for Vet community as well.
Excellent work sir je.proud to be your student
My question is that it is more virulent or less virulent than Wuhan or Network strain we see more deaths of Pakistani born than in our country